-
PROVIDERS
New MRD Medicare Coverage for Select Indications*
*When coverage criteria are met. Additional criteria and exceptions for coverage may apply.
-
LIFE SCIENCES
Register now
UPCOMING WEBINARTranslating data into an actionable R&D strategy
-
PATIENTS
It's About Time
View the Tempus vision.
- RESOURCES
-
ABOUT US
View Job Postings
We’re looking for people who can change the world.
- INVESTORS
09/23/2025
Solving cancer’s tissue scarcity problem: How Tempus uses AI models in pathology to transform tissue analysis
Tempus AI research demonstrates how AI can analyze digitized pathology images to predict tissue sample viability for genomic testing, a task that pathologists struggle to perform consistently.
Authors
Nike Beaubier, MD
SVP of Life Sciences and Pathology, Tempus

Jason Blue-Smith,
VP & General Manager, Algos,
Tempus

SVP of Life Sciences and Pathology, Tempus

Jason Blue-Smith,
VP & General Manager, Algos,
Tempus

In this article we will take a closer look at Paige Predict’s tissue optimization solution, which accurately predicts the likelihood of a sample failing Tempus xT sequencing with 96% specificity.1 Paige Predict also enables significant tissue conservation by identifying samples with a higher TNA [total nucleic acid] yield than is typical, allowing fewer slides from that specimen to be used for NGS testing and ensuring more tissue is preserved for other critical downstream biomarker tests.
The hidden bottleneck in cancer care
Every day, thousands of cancer patients wait anxiously for test results that could determine their treatment plan. What they may not know is that some of these critical genomic tests fail – not because of faulty equipment or human error, but because their tissue samples simply don’t contain enough genetic material to analyze.
These “quantity-not-sufficient” (QNS) failures create a clinically challenging cascade: patients endure additional invasive procedures and treatment decisions can be delayed by weeks, resulting in wasted time and resources. Meanwhile, pathologists face a challenging task: determining whether a microscopic tissue sample contains enough DNA and RNA for next-generation sequencing (NGS), a decision that carries significant implications for patient care, yet often must be made based on visual evaluation alone.
This is where artificial intelligence can be a helping partner. At Tempus, we’ve developed Paige Predict, an AI system that analyzes digitized images of H&E-stained pathology slides to predict nucleic acid yield with unprecedented accuracy. Integrated directly into our laboratory workflow, Paige Predict has transformed one of oncology’s most persistent bottlenecks into a source of intelligent decision-making.
This article explores how this technology was developed and how it is being used to modernize a fundamental step in the cancer care continuum, helping more patients get the answers they need, faster.
Why traditional methods fall short
Cancer diagnosis in the genomic era depends on extracting maximum information from minimal tissue. When a patient undergoes a biopsy, that precious sample must serve multiple purposes: initial diagnosis, molecular profiling for targeted therapy selection, and assessing clinical trial eligibility.
At the center of this process sits the pathologist, examining tissue under a microscope to make critical decisions. Beyond rendering a diagnosis, they must visually estimate how much tissue to allocate for different tests—a process that relies heavily on hard earned experience and intuition.
Traditionally, this estimation relies on visual assessment, a standard practice that is subject to interobserver variability. An inaccurate estimate can have significant consequences. Using too little tissue leads to a QNS failure, which can delay a patient’s access to critical genomic information by weeks. Using too much tissue depletes a finite and precious resource that could be used for other essential biomarker tests.
As the demand for comprehensive molecular profiling grows, these traditional methods struggle to keep pace. AI provides the critical solution, offering a way to standardize and optimize this process by extracting actionable intelligence from pathology images with a level of precision and consistency that complements human expertise.
Tempus’ integrated AI philosophy
At Tempus, we view AI not as a standalone tool, but as a core component of an integrated system that connects data, technology, and medicine. Our approach is built on one of the world’s largest libraries of de-identified, multimodal clinical and molecular data. This vast repository provides the foundation for training and validating sophisticated AI algorithms that can recognize subtle patterns imperceptible to the human eye.
The Paige Predict model was developed using this philosophy. It is not just an algorithm; it is a feature embedded directly into our digital pathology review platform and laboratory developed tests, providing real-time decision support for our laboratory teams. This human-in-the-loop system augments the pathologist’s expertise, providing a quantitative data point to help guide their assessment.
This integrated philosophy is built on four interconnected pillars:
- Speed: Reducing the time from sample to insight.
- Efficiency: Optimizing resources and streamlining workflows.
- Accuracy: Improving the reliability of processes and predictions.
- Data Insights: Uncovering novel patterns to drive discovery.
By focusing on these pillars, our AI systems work together to create a more intelligent and effective pathway for cancer research and care.
Demo: The Paige Predict workflow in action
This video demonstrates the AI-generated yield that appears in the pathologist’s digital review interface, enabling informed decision-making to prepare for molecular profiling.
How Paige Predict transforms the laboratory workflow
The Paige Predict algorithm exemplifies how AI can deliver tangible improvements across our four pillars. By analyzing a digitized image of a standard H&E-stained slide, the model provides crucial predictions that optimize the entire NGS workflow.
- Accelerating with speed: In a traditional workflow, a QNS failure is often discovered only after the sequencing process is complete. Paige Predict helps to avoid this delay. The model delivers its prediction instantly within the pathologist’s digital review tool, flagging samples at high risk of failure for human review before they ever enter the sequencing pipeline. This speed allows our teams to take immediate action, such as requesting additional tissue from the provider. By turning a potential two-week delay into a near real-time intervention, we accelerate the timeline for delivering critical results to physicians and their patients.
- Driving efficiency: Paige Predict automates and standardizes the yield estimation process, reducing manual variability and optimizing the use of every tissue sample. The model provides a data-driven recommendation for the number of tissue sections to scrape, helping to prevent the two most common inefficiencies:
-
- Under-sampling: By identifying samples likely to fail, Paige Predict reduces the significant laboratory costs and resources wasted on unsuccessful sequencing runs.
- Over-sampling: The model also identifies cases expected to yield an excess of TNA. This allows pathologists to conserve tissue, preserving it for other vital tests, such as immunohistochemistry (IHC). This efficiency ensures that we maximize the clinical utility of every nanogram of tissue.
- Enhancing accuracy: The primary function of Paige Predict is to accurately predict TNA yield and, consequently, the likelihood of a sample failing Tempus xT sequencing. The model was trained and validated on hundreds of thousands of real-world cases from the Tempus database. In our validation studies, the model demonstrated robust performance across multiple cancer and procedure types, achieving a high degree of specificity in predicting both low-yield samples and those that would fail sequencing. This accuracy provides our pathologists with a reliable tool to support their decisions, ultimately reducing the overall QNS rate and increasing the number of patients who successfully receive a comprehensive molecular report.
- Unlocking data insights: By learning from an immense dataset of pathology images and their corresponding sequencing outcomes, Paige Predict uncovers complex morphological patterns that correlate with nucleic acid yield. These insights extend beyond what is visible to the human eye. The model provides a deeper understanding of how factors like tumor type, cellularity, and tissue architecture impact the quality of a sample. This continuous feedback loop not only improves the model itself but also generates broader insights that can inform our entire laboratory process, helping us understand which samples are best suited for which assays and driving a more intelligent approach to testing.
The science behind the breakthrough
The development and performance of our AI models are validated through rigorous scientific research and shared transparently with the clinical community.
From slide to prediction: The AI pipeline
The Paige Predict algorithm transforms a standard pathology slide into a quantitative prediction through a multi-step process.
- Digitization: The process begins when a physical H&E-stained glass slide is scanned to create a high-resolution whole slide image (WSI).
- AI-Powered Cell Segmentation: The WSI is then analyzed by an AI model based on a U-Net architecture. This specialized network examines the entire image and performs cell segmentation, identifying and counting every cell present in the tissue. Initial development leveraged comprehensive tissue classification models that extracted imaging features including total cell count, lymphocyte count, tumor shape, cell shape, and cell color/texture properties. Total cell count alone carried over 90% of the predictive signal when correlated with ground truth TNA yield.2
- Feature Extraction and Data Integration: To achieve pan-cancer generalizability, the system relied on regularized linear regression and was refined to focus on the most robust features: total cell count (which performs consistently across all cancer types) and a restricted set of clinical metadata known to impact yield, including procedure type (e.g., biopsy, resection), tissue site, sample age, and source institution. This streamlined approach supported reliable performance across diverse cancer types while maintaining predictive accuracy.2
- Yield Prediction: Rather than relying solely on a direct linear regression on features, the final model employs a systematically constructed algebraic expression with decay constants to better capture the expected sub-linear relationships between features and predicted TNA yield. This expression was optimized by measuring impact on AUC for predicting TNA mass < 100ng, with coefficients subsequently calibrated to optimize QNS rate predictions. The model generates two key outputs: the likelihood of the sample failing DNA sequencing due to insufficient quantity (DNA QNS) and estimates for the number of slides needed to obtain sufficient yield.2
Infographic: How Paige Predict analyzes a sample
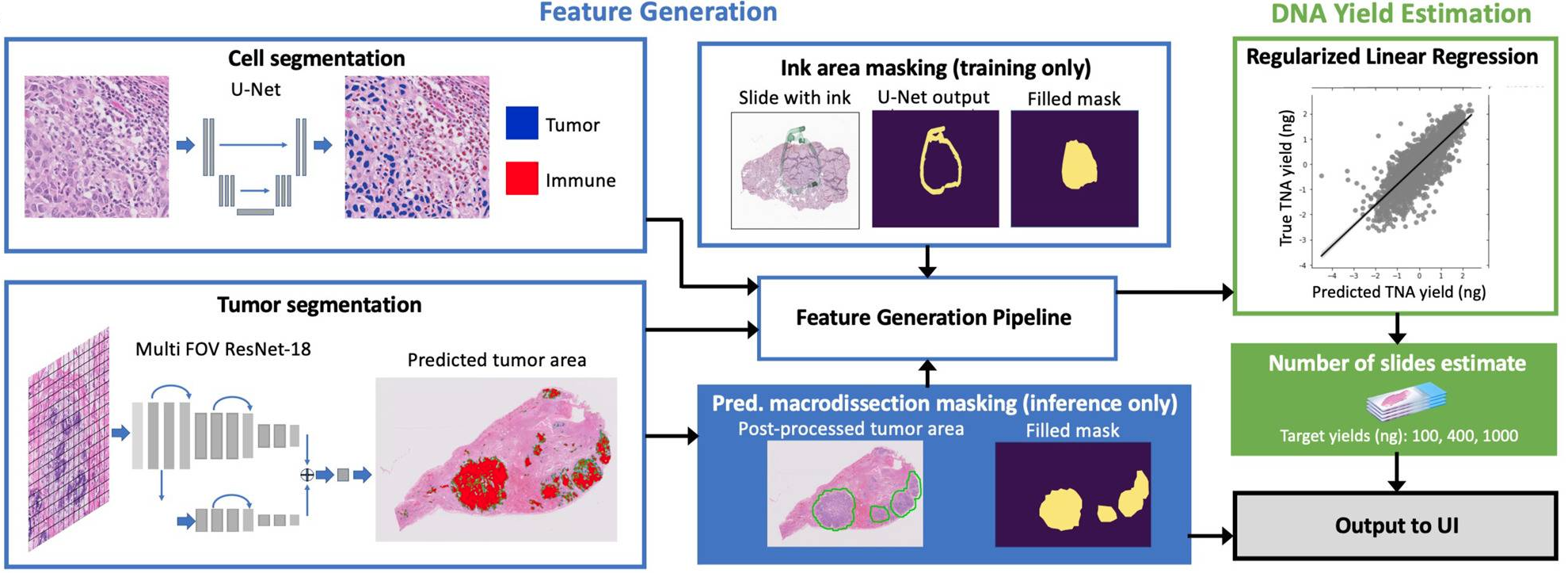
This infographic illustrates the digital pathology workflow with Paige Predict. It shows how an H&E slide image and clinical metadata are processed by the AI model to generate a predictive yield score, which helps optimize the sample for next-generation sequencing.
Rigorous validation establishes clinical value
Paige Predict was trained and then validated on a temporally separate set of tens of thousands of real-world samples, ensuring the model’s performance was tested on new data it had never seen before. The results demonstrate its accuracy and reliability.
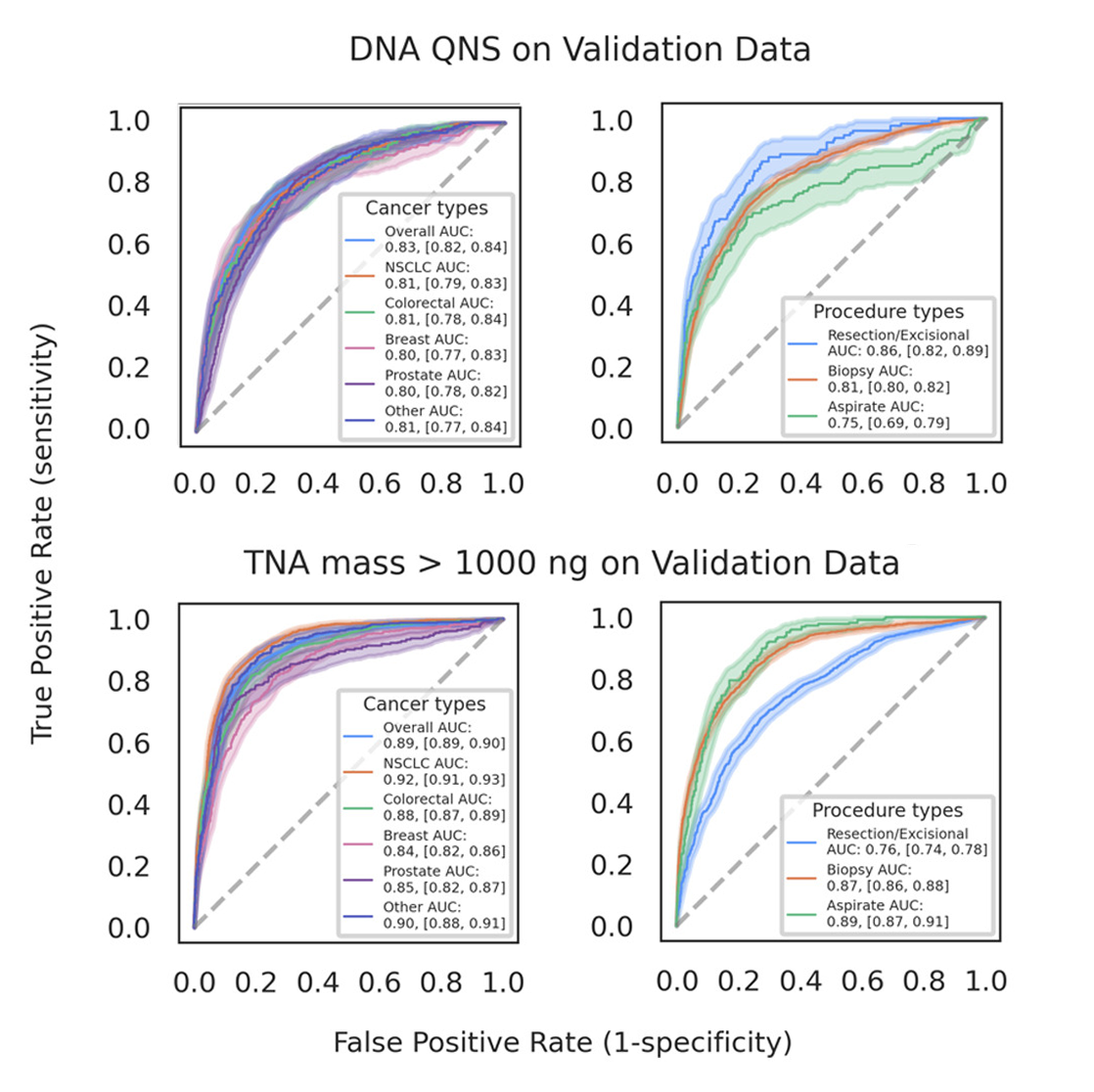
High specificity means you can trust the results. Across the validation set, Paige Predict demonstrated the likelihood of a sample failing Tempus xT sequencing with 96% specificity.1
- What this means: The model has a very low false alarm rate. When Paige Predict does not flag a sample, you can proceed with confidence, knowing there is a high probability that the sample will yield sufficient material for sequencing. This prevents unnecessary workflow disruptions for samples that are viable.
Strong positive predictive value (PPV) means flags are meaningful. When Paige Predict does identify a sample as high-risk, the prediction is highly reliable.
- The proof: In our validation studies, samples flagged by the model as likely to fail either DNA or RNA sequencing had a PPV of 80%. This means that 8 out of 10 samples flagged for this reason did, in fact, fail. This high degree of accuracy demonstrates that when an alert is raised, it warrants attention and intervention.1
Robust performance across diverse cancer and sample types. The model’s accuracy is not just an overall average; it remains strong across the most common cancer and procedure types, proving its utility in pathologists’ day-to-day work.
- The proof: When predicting low-yield samples, the model’s PPV was consistently high across major cancer types, including 81% in non-small cell lung cancer (NSCLC), 80% in prostate cancer, and 86% in breast cancer. Whether it is used with a core biopsy or a fine-needle aspirate, the model provides a reliable prediction.1
Proven impact on tissue conservation. Beyond preventing failures, Paige Predict is also highly effective at identifying samples with abundant TNA, enabling good tissue stewardship. The model predicted samples yielding over 1,000 ng of TNA with 73% sensitivity and 89% specificity.
- What this means: Pathologists can confidently identify cases where fewer tissue sections are needed for sequencing. This preserves precious and finite tissue for other critical assays, such as IHC or other biomarker tests, maximizing the diagnostic information gained from a single patient sample.
This scientific validation, published and presented at leading oncology conferences like the United States and Canadian Academy of Pathology (USCAP), confirms that Paige Predict is a reliable, evidence-based tool designed to bring greater precision and confidence to the pathology workflow.
Receiver operating characteristic curves for labels stratified by clinical variables
Patient case: Lung adenocarcinoma with low tumor purity
Tempus has implemented Paige Predict into our pathology workflow and LIMS system for use with our laboratory developed tests, which allows pathologists to review cases remotely, select areas for macrodissection, and select the optimal number of slides for DNA/RNA isolation.
An example of this AI in clinical use came from a patient with lung adenocarcinoma. The tumor was immune infiltrated and had a considerable amount of reactive stroma with the benign lung area containing numerous alveolar macrophages. This led to a lower tumor content (percent tumor versus normal cells) – as predicted by the algorithm – than ideal for sequencing. We used Paige Predict in combination with macrodissection (shown below) to enrich the tumor content and select the correct number of slides to obtain a sufficient quantity of nucleic acid. This approach led to successful NGS sequencing using only five slides, thus preserving tissue for subsequent testing including IHCs.
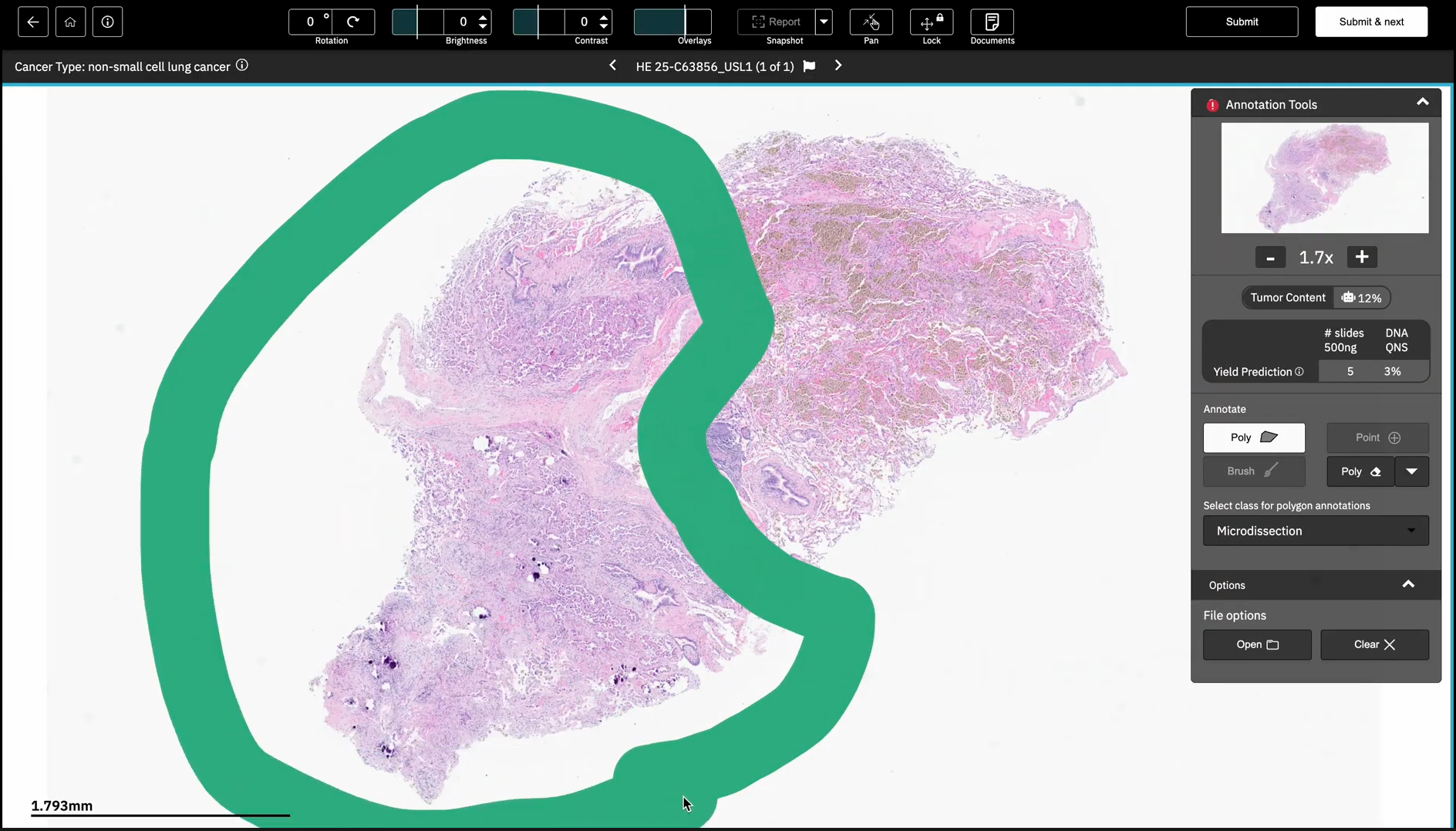
Tempus pathologists can indicate areas for macrodissection and see the Paige Predict predictions within a digital pathology viewer.
Transforming cancer care, one sample at a time
Tempus is leveraging AI to solve fundamental challenges in oncology, and Paige Predict is a clear demonstration of that strategy in practice. By focusing on speed, efficiency, accuracy, and data insights, this tool transforms a critical laboratory step into a source of intelligence.
We are partnering with select pathology laboratories to offer Paige Predict to early research collaborators. Through this beta program, we will integrate with external lab systems to explore the potential of Paige Predict’s predictive analytical capabilities. This collaboration with the broader oncology community is an important step in understanding how AI-driven tools can support future advancements in digital pathology.
The future of oncology is one where human expertise and artificial intelligence work together, setting new standards for precision and efficiency in patient care. By augmenting the expertise of pathologists with powerful AI, we are helping to ensure that more patients can benefit from the full potential of precision oncology.
To learn more about how we can innovate pathology together, contact us today.
1. Ingale K, Och J, Nagpal K, et al. Validation and deployment of H&E image based model predicting total nucleic acid yield in multiple cancer types. Lab Invest. 2024;105(3):103598
2. Osinski BL, BenTaieb A, Ho I, et al. Artificial intelligence-augmented histopathologic review using image analysis to optimize DNA yield from formalin-fixed paraffin-embedded slides. Mod Pathol. 2022;35(12):1791-1803. doi:10.1038/s41379-022-01161-0
-
01/20/2026
From image to insight: How Tempus is leveraging AI to advance biomarker screening at scale
By leveraging Paige Predict to screen for genomic and phenotypic biomarkers, clinicians and researchers have rapid access to valuable information to prioritize testing and pre-screen patients at scale.
Read more -
12/04/2025
Pioneering decentralized oncology trials: Success at the nation’s largest community practices
Hear directly from industry leaders and pioneering site partners as they share insights on expanding their research footprint and improving financial sustainability for their institutions.
Watch replay -
11/06/2025
Beyond imaging: Leveraging molecular tools to detect MRD for personalized cancer care
This webinar explores the clinical value of MRD testing and IO treatment response monitoring in oncology, using real-world cases to illustrate its application in patient management.
Watch replay


