-
PROVIDERS
Watch now
Are you getting the full picture? A webinar series on the power of comprehensive intelligent diagnostics
-
LIFE SCIENCES
REGISTER NOW
Closing Care Gaps with AI: The Next Competitive Edge in Pharma
Monday, July 14
9am PT, 11am CT, 12pm ET -
PATIENTS
It's About Time
View the Tempus vision.
- RESOURCES
-
ABOUT US
View Job Postings
We’re looking for people who can change the world.
- INVESTORS
03/25/2024
Large-Scale DNA Alteration Prediction From H&E Images Across Different Cancer Types Generalizing To External Images
USCAP 2024 Annual Meeting
PRESENTATION
Authors
Kshitij Ingale, Sun Hae Hong, Boleslaw L Osinski, Mina khoshdeli, Josh Och, Rohan P Joshi, Kunal Nagpal, Martin C Stumpe
Background: Molecular alterations detected by Next Generation Sequencing (NGS) have predictive, prognostic, and diagnostic value across various cancer types. NGS may sometimes not be ordered in patients who may benefit due to low pretest probability, tissue availability, and cost considerations. Algorithms that use digitized H&E stained images routinely acquired in the clinical workflow to rapidly screen patients for molecular alterations could be used to prioritize patients for NGS testing. In this study, we identified a broad range of molecular alterations that can be detected using H&E images and evaluated the generalizability of a subset of these models on cases obtained from external labs and from The Cancer Genome Atlas (TCGA) projects.
Design: For this multi-cancer type study, a dataset of 2928 bladder cancer (3652 for FGFR), 2949 endometrial cancer, and 13,061 colorectal cancer whole slide images along with corresponding DNA alteration labels obtained from NGS was curated. Models were trained to predict the mutation from H&E-stained images. Additionally, logistic regression models with clinical and operational covariate features were trained as a reference. We consider an imaging model to have a signal if it outperforms the reference model based on effect size (ROC-AUC improvement by 5%) and statistical significance (FDR corrected p > 0.05). A subset of actionable molecular target models was evaluated on a test set consisting of slides stained at external labs as well as a test set with slides sourced from TCGA projects.
Results: Out of 78 imaging models, 46 (59%) outperformed corresponding reference models as shown in Figure 1, suggesting an association of morphological features with many DNA alterations. The model performance is stable on data obtained from external labs with a mean drop of 0.01 in ROC-AUC (Figure 2a). The data from TCGA projects is obtained from a diverse set of external labs and clinical characteristics, however, the models appear to be reasonably robust to these cases as well, resulting in a mean drop of 0.04 in ROC-AUC (Figure 2b).
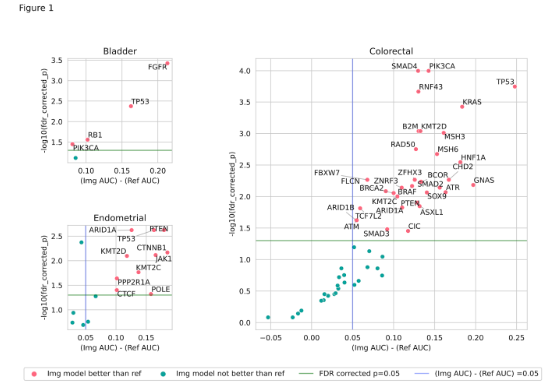
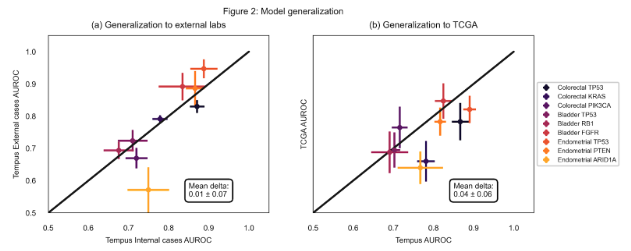
Conclusion: Our work suggests that H&E image-based identification of actionable molecular mutations can be a
viable strategy for patient prioritization. Some of the actionable target algorithms were also generalizing to external demonstrating potential for a real-world deployment. Further rigorous model development can be explored for integrating these models into clinical practice.