-
PROVIDERS
Watch now
Are you getting the full picture? A webinar series on the power of comprehensive intelligent diagnostics
-
LIFE SCIENCES
REGISTER NOW
Closing Care Gaps with AI: The Next Competitive Edge in Pharma
Monday, July 14
9am PT, 11am CT, 12pm ET -
PATIENTS
It's About Time
View the Tempus vision.
- RESOURCES
-
ABOUT US
View Job Postings
We’re looking for people who can change the world.
- INVESTORS
03/25/2024
Pan-Cancer Nuclei Segmentation in Hematoxylin And Eosin Whole Slide Images
USCAP 2024 Annual Meeting
PRESENTATION
Authors
Mina Khoshdeli, Rohan Joshi, Bolesław L. Osinski, Luca Lonini, Martin C. Stumpe
Background
Nuclei segmentation is a critical stage in characterizing the morphology of cells in Hematoxylin and Eosin (H&E) stained whole slide images (WSIs). Extensive research has been conducted on the application of deep learning models for nuclei segmentation. While individual models have shown promising performance in segmenting nuclei for specific cancer types, a gap remains in the availability of a single model to segment nuclei across cancer types. Here, we created a comprehensive training and validation dataset encompassing a broad spectrum of cancer types and histological subtypes to address this need.
Design
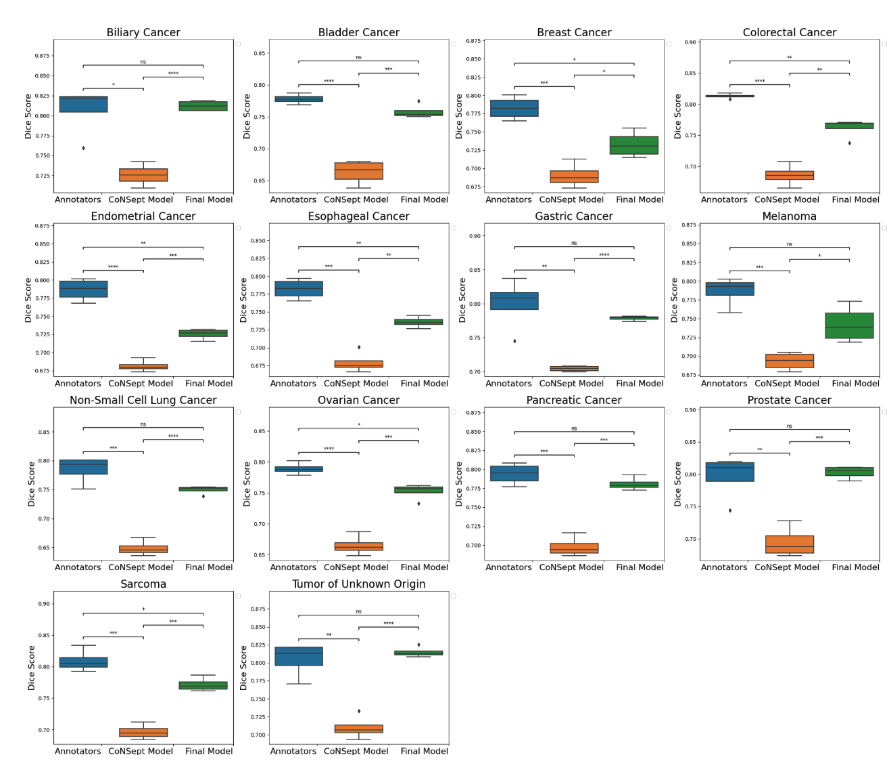
A cohort of WSIs was generated including 14 cancer types, with each having up to three histological subtypes (N=410, Table I). From these WSIs, we randomly selected two fields of view (FOV) from the tissue area for annotation at 40x magnification. During the training phase, we acquired one annotation per image, while for the validation phase, we gathered four annotations per image. We trained a HoverNet model initializing using weights from a model previously trained on the CoNSeP dataset. To assess the model performance, we conducted a comparative analysis between the trained model, the initial CoNSeP-trained model, and consensus evaluations of pathologists. To determine consensus among pathologists, we computed majority votes for all possible permutations of three pathologists, with one annotator being excluded for each permutation. We compared the consensus annotations with the model’s output and the annotations of the held-out pathologists. As a result, we obtained four data points for the box plots presented in Figure1 and 2. Statistical testing was performed using a t-test for independent samples and Bonferroni correction.
Results
We used a cell-based dice score to compare the performance of models and human annotators. We found comparable performance between the model’s output and the performance of human annotators (mean 0.767, 95% CI 0.760-0.773 for the model; mean 0.794, 95% CI 0.777-0.810 for human annotators, p=0.08 with Bonferroni correction). In prostate, non-small cell lung, pancreatic, tumor of unknown origin, melanoma, gastric, bladder, and biliary cancers we found statistically indistinguishable performance in subgroup analyses.
Conclusions
In this study, we have established an annotation dataset tailored explicitly to pan-cancer nuclei segmentation and have demonstrated that by utilizing the HoverNet architecture, we have achieved near-human-level performance.
Table 1
| Cancer Type | Histology subtypes | Number of images in Training | Number of images validation |
| Biliary Cancer | cholangiocarcinoma | 12 | 12 |
| Bladder Cancer | urothelial carcinoma | 12 | 12 |
| Breast Cancer | breast carcinoma | 10 | 11 |
| Colorectal Cancer | colorectal adenocarcinoma | 12 | 12 |
| Endometrial Cancer | carcinosarcoma | 12 | 12 |
| endometrial serous carcinoma | 12 | 12 | |
| endometrioid carcinoma | 11 | 12 | |
| Esophageal Cancer | gastroesophageal adenocarcinoma | 12 | 12 |
| gastroesophageal squamous cell carcinoma | 12 | 12 | |
| gastroesophageal adenocarcinoma | 12 | 12 | |
| Melanoma | melanoma | 12 | 12 |
| Non-Small Cell Lung Cancer | lung adenocarcinoma | 12 | 12 |
| lung squamous cell carcinoma | 12 | 12 | |
| Ovarian Cancer | ovarian serous carcinoma | 12 | 11 |
| Pancreatic Cancer | pancreatic adenocarcinoma | 12 | 12 |
| pancreatic neuroendocrine tumor | 10 | 12 | |
| Prostate Cancer | prostatic adenocarcinoma | 12 | 12 |
| Sarcoma | fibrous sarcoma | 11 | 12 |
| leiomyosarcoma | 12 | 12 | |
| Tumor of Unknown Origin | NA | 36 | 36 |
Figure 1
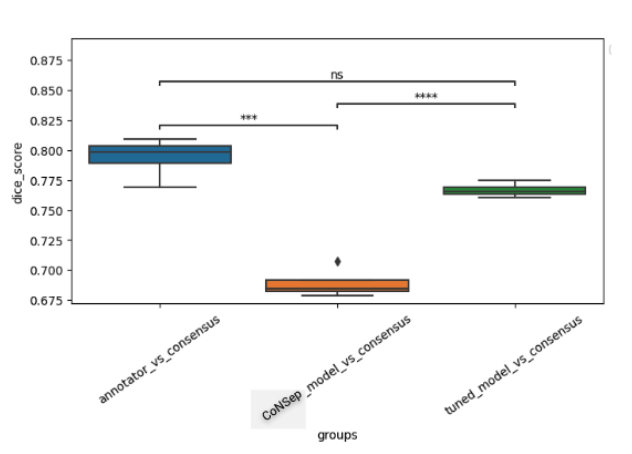
Figure 2