-
PROVIDERS
REGISTER NOW
Navigating new frontiers in breast cancer care pathway intelligence: The role of providers and AI
Tuesday, July 29th
2:00pm PT, 4:00pm CT, 5:00pm ET -
LIFE SCIENCES
REGISTER NOW
Navigating New Frontiers in Breast Cancer Care Pathway Intelligence: The Role of Providers and AI
Tuesday, July 29
2pm PT, 4pm CT, 5pm ET -
PATIENTS
It's About Time
View the Tempus vision.
- RESOURCES
-
ABOUT US
View Job Postings
We’re looking for people who can change the world.
- INVESTORS
04/29/2024
Using an AI algorithm to predict possible MSI high status in prostate cancer patients from digital pathology images
Tissue sequencing in prostate cancer is often challenging because of limitations of tumor quantity or tissue quality; bone metastases are frequent. It is critical to identify whether patients are MSI-H since they may be eligible for immunotherapy and/or may need workup for Lynch syndrome.
Authors
Nike Beaubier, MD
SVP of Life Sciences and Pathology, Tempus

SVP of Life Sciences and Pathology, Tempus

With our ability to digitize pathology slides and use AI to garner insights from these images, we can support efficient use of tissue while also helping to surface appropriate patients for dMMR/MSI-H testing to ensure guideline-directed care for those with relevant alterations.
Nike Beaubier, MD and SVP of Life Sciences and Pathology shares her perspective on these issues and how Tempus’ newly launched p-MSI algorithm has helped predict MSI-H status for prostate cancer patients who otherwise may not have received MSI results.
Combining AI algorithms and digital pathology can streamline the diagnosis and treatment of prostate cancer patients
Tissue sequencing in prostate cancer is challenging because tumor content is often low, and metastases are predominantly in the bone, particularly in the later stages of disease. The prevalence of the tissue-agnostic biomarker microsatellite instability-high (MSI-H) / deficiency in mismatch repair (dMMR) in prostate cancer is approximately 2-3%1,2.
Many prostate cancer patients do not currently receive MSI status results from NGS sequencing because of sample quantity or quality issues. The tumor fraction must be ≥ 30% on a successful sequencing run for the NGS MSI-H algorithm to produce a result.
It is critical to identify whether patients are MSI-H in metastatic prostate cancer, since this patient population may be eligible for immunotherapy. Response rates as high as 50% have been observed when dMMR/MSI-H prostate cancer patients are treated with immune checkpoint inhibitors (ICI), making the case for MSI-H testing as a gateway to these patients accessing beneficial therapies.2
Early-stage prostate cancer patients can also benefit from knowing their MSI status because many dMMR/MSI-H cases are due to Lynch syndrome, so they and their families may benefit from genetic testing and cancer screening.
“At Tempus we’ve been sequencing patients for years, so we have rich matched molecular data—including RNA whole transcriptome data—and hundreds of thousands of whole slide images. Combining de-identified data from an imaging library and a molecular library enables Tempus to create AI-driven models that can provide insights which would be unimaginable in any other way.” – Nike Beaubier, MD and SVP of Life Sciences and Pathology
Tempus’ p-MSI algorithm predicts patients with prostate cancer who may be MSI-H
Tempus has developed p-MSI, a novel digital pathology algorithm to surface prostate cancer patients who are more likely than average to be MSI-H and who should be considered for confirmatory testing.
The p-MSI algorithm runs on digital images of hematoxylin and eosin (H&E) stained slides and therefore does not consume tissue to run since the H&E is already required for diagnosis. Upon scanning the H&E, the algorithm predicts whether a patient is more likely than the typical prostate cancer patient to be MSI-H/dMMR. Then, informed by this prediction, the treating oncologist or urologist can opt to do confirmatory testing such as immunohistochemistry (IHC) staining for MMR proteins. If IHC is selected as the modality for confirmatory testing, as few as 100 cells/slide can yield a result—significantly below what is required for next-generation sequencing (NGS).
By identifying an enriched population of prostate cancer patients who are more likely to be MSI-H, Tempus is helping to direct patients towards targeted treatments.
The mismatch repair protein IHC panel (MLH1, PMS2, MSH2, MSH6) is widely available, supporting broad access to confirmatory testing. As a result, the insight gained from running p-MSI on patient samples isn’t just nice to have, it’s actionable.
The synergy between digital pathology and AI can transform the future of precision medicine
Although the human eye and brain are remarkable at analyzing images, AI algorithms can detect patterns the human eye does not. Recently, AI has proven beneficial in analyzing text and deriving insights from textual data. Images are far more data dense than text, meaning AI has a rich data source to work with when analyzing images for deeper patterns and insights. Furthermore, AI has a significantly higher processing capacity than the human brain and can analyze hundreds of thousands of features on a slide and tens of thousands of slides at a time.
AI can not take the place of an expert pathologist; rather, it boosts their efficiency and accuracy, by offering insights beyond the ability of human visual analysis. Together, we believe pathologists and AI achieve outcomes superior to what either could do alone.
Practically speaking, Tempus has a unique infrastructure to leverage AI algorithms within digital pathology to augment clinical practice. Tempus has been sequencing patients for years and has always scanned slides, so we have a rich library of matched molecular data—including RNA whole transcriptome data—and hundreds of thousands of whole slide images. Using these de-identified molecular datasets, the research team can train imaging models from molecular labels, and the algorithms can look for imaging features across hundreds of thousands of cases to recognize patterns. Combining de-identified data from an imaging library and a molecular library enables Tempus to create AI-driven models that can provide insights which would be unimaginable in any other way.
Using AI algorithms alongside digital pathology will help catalyze advances in precision medicine that will improve clinical practice over time. Radiology workflows have been digitized for many years, with images of tissue captured directly by CCD cameras, to great benefit. However, pathology images can not be captured directly from tissue; tissue must be fixed, processed, and stained on glass slides first. Thus, digital image capture has not universally been done at large scale. However, the advancement of AI algorithms, like the one described here, provides the incentive to do so. Pathologists and other patient care providers can generate insights using these digital images and bring patients closer to receiving the treatments they urgently need.
1. Hu, Q., Rizvi, A. A., Schau, G., Ingale, K., Muller, Y., Baits, R., … & Nagpal, K. (2023). Development and Validation of a Deep Learning-Based Microsatellite Instability Predictor from Prostate Cancer Whole-Slide Images. arXiv preprint arXiv:2310.08743.
2. Abida W, Cheng ML, Armenia J, Middha S, Autio KA, Vargas HA, Rathkopf D, Morris MJ, Danila DC, Slovin SF, Carbone E, Barnett ES, Hullings M, Hechtman JF, Zehir A, Shia J, Jonsson P, Stadler ZK, Srinivasan P, Laudone VP, Reuter V, Wolchok JD, Socci ND, Taylor BS, Berger MF, Kantoff PW, Sawyers CL, Schultz N, Solit DB, Gopalan A, Scher HI. Analysis of the Prevalence of Microsatellite Instability in Prostate Cancer and Response to Immune Checkpoint Blockade. JAMA Oncol. 2019 Apr 1;5(4):471-478. doi: 10.1001/jamaoncol.2018.5801. PMID: 30589920; PMCID: PMC6459218.
-
07/01/2025
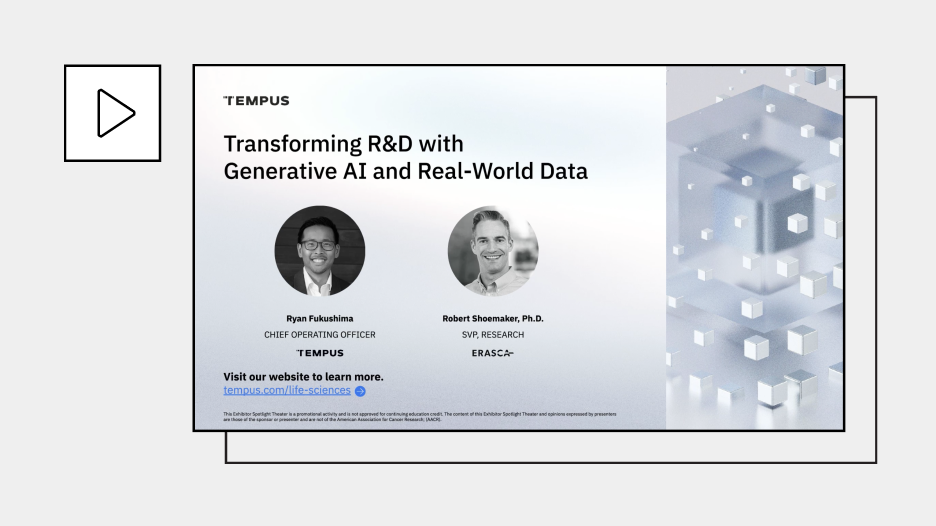
Transforming R&D with Generative AI and Real-World Data
Mike Yasiejko, General Manager and Executive Vice President at Tempus, recently spoke with Andrew Mazar, PhD, Chief Operating Officer at Actuate Therapeutics, about the ways Tempus’ liquid biopsy (xF+) and methylation assays have been valuable to Actuate’s work and how the company has benefitted from the partnership
Watch now -
06/12/2025
AI & ML in action: Demonstrating real-world impact in trial design & patient care
Discover how the Tempus platform leverages AI and ML to inform standard of care practices through health equity guidelines and drive insights that help refine clinical trial design. Engage with live demonstrations showcasing how our tools identify patients by modifying inclusion/exclusion criteria and leveraging patient queries. Explore how our tools integrate NCCN guidelines and empower life science teams to access current, actionable patient-journey insights. Learn how these real-world applications can drive progress in your clinical development initiatives.
Watch replay
Secure your recording now. -
06/09/2025
Bridging the translational gap: The role of organoids in oncology R&D
This white paper explores the evolving role of organoids in oncology R&D, highlighting their potential as predictive preclinical models and their ability to reduce translational risk. Download for a comprehensive overview of the scientific landscape, key adoption barriers, emerging innovations, and how pharma companies leverage organoids to accelerate precision medicine.
Read more