-
PROVIDERS
Watch now
Are you getting the full picture? A webinar series on the power of comprehensive intelligent diagnostics
-
LIFE SCIENCES
REGISTER NOW
Closing Care Gaps with AI: The Next Competitive Edge in Pharma
Monday, July 14
9am PT, 11am CT, 12pm ET -
PATIENTS
It's About Time
View the Tempus vision.
- RESOURCES
-
ABOUT US
View Job Postings
We’re looking for people who can change the world.
- INVESTORS
03/25/2024
H&E-Based Deep Learning Model Predicts Immune Phenotypes in NSCLC
USCAP 2024 Annual Meeting
PRESENTATION
Authors
Josh Och, M.S., Bolesław Osinski, Ph.D., Sina Zomorrodian, Rohan Joshi, Nike Beaubier, Martin C. Stumpe
Background – Tumor-infiltrating lymphocytes (TIL) are a biomarker for response to immune checkpoint inhibitor (ICI) therapy. However, visually identifying TILs from hematoxylin and eosin (H&E) stained whole-slide images (WSIs) is expensive and time-consuming. Further, a lack of consensus on TIL scoring criteria can result in high levels of pathologist discordance. Here, we use an imaging-based deep learning model to classify H&E WSIs of non-small cell lung cancer (NSCLC) tumors into inflamed (TIL infiltrated) versus non-inflamed immune phenotypes.
Design – H&E stained WSIs (scanned on Leica Aperio GT450) from primary site, resected NSCLC tumors enriched for class balance between inflamed and non-inflamed (n=169, Table I) were assessed by 4 independent pathologists who assigned one of the following labels: inflamed, excluded (stromal TIL infiltrated) or desert (no infiltration). WSIs were randomly split (50:50) into training and testing sets. For model training, one pathologist was held out; each WSI was assigned a ground truth label if 2/3 of remaining pathologists agreed. Previously developed deep learning models were used to count the number of lymphocytes in tumor and stroma regions in 1 mm2 tiles. WSIs were called “inflamed” if enough tiles (slide threshold) had a sufficient number of lymphocytes within tumor regions (tile threshold). Tile and slide-level thresholds were tuned to maximize the F1 score for separation of inflamed and non-inflamed (either excluded or desert) cases in the training set. Model performance was assessed on the independent held-out test set and compared to calls from the held-out pathologist. This process was repeated 4 times, holding out each pathologist.
Results – Overall, our deep learning model achieved a mean of 77.5% accuracy (95% CI 73.3% – 81.7%), which was indistinguishable from that of the held-out pathologists (mean 80.3%, 95% CI 69.0% – 91.6%). Cohen’s kappa coefficient for the model (mean 45.4%, 95% CI 34.9% – 55.9%) was also indistinguishable from that of the held-out pathologists (mean 49.2%, 95% CI 25.3% – 73.0%). A representative comparison of the model to a held out pathologist is shown in Fig.1.
Conclusions – Our deep learning model performed as well as a held-out pathologist for separating inflamed vs. non-inflamed samples. Future work includes extending our model to predicting excluded vs. desert phenotypes, identifying tertiary lymphoid structures, and validating that model predictions are relevant for therapy response.
Figure 1
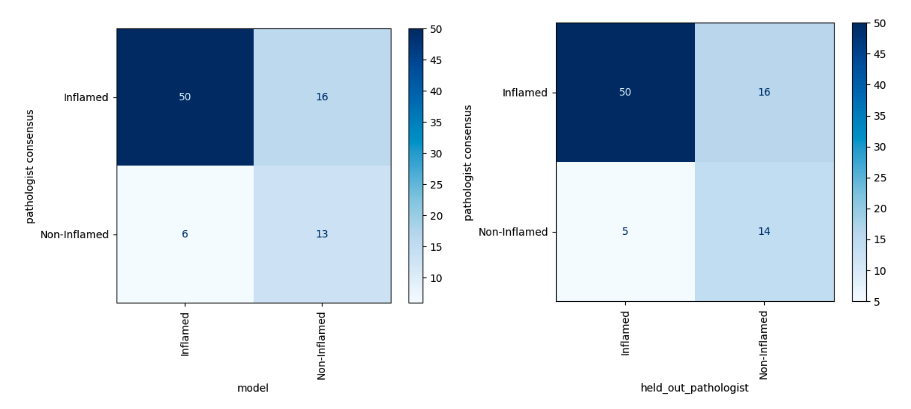
Table 1
| Characteristic | Value | Samples | Percent Inflamed (defined by majority of three pathologists) | Chi-square p-value |
| Stage | Stage 1 | 48 | 77% | 0.018 |
| Stage 2 | 54 | 85% | ||
| Stage 3 | 50 | 64% | ||
| Stage 4 | 17 | 53% | ||
| Grade Rollup | Low Grade | 105 | 70% | 0.204 |
| High Grade | 64 | 80% | ||
| Histology | Adenocarcinoma | 86 | 71% | 0.681 |
| Acinar Cell Carcinoma | 50 | 74% | ||
| Mucinous Adenocarcinoma | 33 | 79% |