-
PROVIDERS
REGISTER NOW
Scientific breakthroughs across the cancer continuum: Research from the 2024 ASCO® Annual Meeting
Join us as we engage in a multi-disciplinary discussion around practice changing approaches in patient management across the cancer care journey. -
LIFE SCIENCES
REGISTER NOW
Integrating omics in R&D: A new era of biomedical research
Join us as industry experts discuss the integration of diverse omics data, from next-generation sequencing (NGS) to proteomics, and explore its transformative impact on the research landscape. -
PATIENTS
It's About Time
View the Tempus vision.
- RESOURCES
-
ABOUT US
View Job Postings
We’re looking for people who can change the world.
- INVESTORS
08/05/2022
Getting to the heart of multimodal data
Go behind the scenes with Tempus COO Ryan Fukushima and learn how harmonized multimodal data sources help accelerate and de-risk drug development programs.
Authors
Ryan Fukushima
COO, Tempus

COO, Tempus

Introduction
For decades, the biopharmaceutical industry has used single sources of real-world data, including claims, electronic health records (EHRs), and cancer registries. Yet no one had amassed a large, harmonized dataset from hundreds of hospitals that included abstracted clinical notes, longitudinal EHR information, tumor and germline DNA sequencing data, whole transcriptome RNA sequencing data, and whole-slide pathology images. As a result, researchers could not accelerate evidence generation for the precision medicine era.
This was the problem Eric Lefkofsky and I set out to solve over seven years ago: could we organize and harmonize heterogeneous data sources at scale to give providers and researchers the tests, data, tools, and insights they need to make precision medicine accessible to patients. Since many underlying questions require a deeper biological understanding, we knew we had to build something that could organize multiple data modalities. We called it “multimodal data,” and we are continuously expanding the Tempus Database with new modalities, including radiology images, electrocardiogram records, and patient-reported outcomes. Today, Tempus is focused on scaling this de-identified dataset to analyze biopharmaceutical companies’ critical research questions – from early drug discovery through clinical development in oncology, cardiovascular disease, and neurology – to help bring better treatment options to patients.
Using our solution, the real-world evidence community can now capture more data on patient populations to understand oncology at a molecular level across those populations. For example, recent research from Tempus makes use of multimodal data to address the question, “How can we better predict a patient’s response to immunotherapies?” Other important applications of heterogeneous data sources include:
- Prioritizing and validating actionable biomarkers or pathway activity
- Designing clinical trials with those biomarkers
- Building evidence on medical product safety and effectiveness
As we continue to build empirical evidence for these applications using heterogeneous data sources, we will identify new, standardized data modalities.
Data types and parameters
Tempus uses multiple important sources of data relevant to drug development. Our current platform includes molecular, clinical, and pathology slide imaging data from real-world care of today. Current examples of molecular data include:
- Tumor/normal DNA sequencing (600+ genes)
- Whole transcriptome RNA sequencing (all genes)
- Immuno-oncology data derived from DNA/RNA
Examples of clinical data include:
- Diagnosis
- Treatments
- Clinical outcomes
Whole-slide pathology imaging data include parameters based on immunohistochemistry (IHC) and hematoxylin and eosin (H&E) assessments.
There are three key parameter considerations for researchers and their stakeholders:
- Usefulness
- Usability
- Completeness
From Tempus’ curation model, to harmonization across EHR feeds, to available delivery of both raw molecular data and analysis-ready flat files, Tempus continues to ensure that its data is usable and useful in comparative analyses. The Tempus Database captures analysis-relevant parameters that rely on the existing data within medical records or clinical notes.
Real world data isn’t perfect and we need to take additional measures to continuously improve data completeness. Tempus harmonizes data from EHR integrations, cultivates relationships with care teams to obtain longitudinal data outside of integrations, and links to a variety of third-party datasets (e.g., claims), all while maintaining controls to protect patient privacy and data security.
Data model
To illustrate the utility of our data model, Tempus emulated the landmark KEYNOTE-024 study, which compared pembrolizumab to chemotherapy for the treatment of PD-L1–positive non-small cell lung cancer (NSCLC). Using the Tempus Database, we selected patients who would have met the enrollment criteria for the study:
- Untreated stage IV NSCLC, ECOG 0-1
- PD-L1 TPS >= 50 percent
- No activating EGFR or ALK alterations
- No untreated brain metastases
- No active autoimmune diseases
Median real-world overall survival estimates for both the chemotherapy- and pembrolizumab-treated cohorts were consistent with those seen in the published study.
Figure 1. Results from Tempus’ trial emulation of KEYNOTE-024
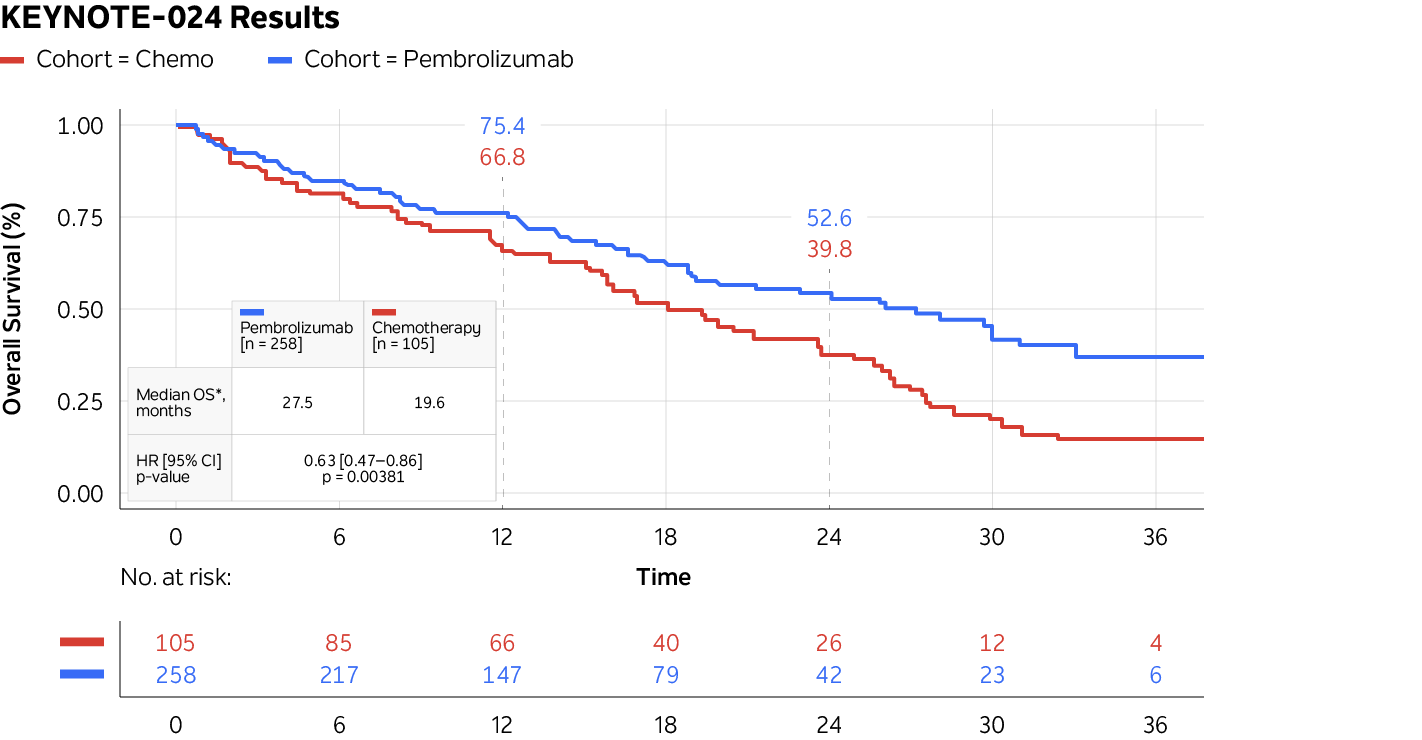
Source: Tempus Database and analysis
Insights
To improve understanding of which subpopulations are more likely to respond to an immunotherapy treatment, Tempus takes a principled approach. We use a bottom-up approach to signature generation; then, we use the Tempus Database to evaluate the outcome. For example, in a recent publication, we performed a research study using single-cell RNA sequencing (RNA-Seq) on tumor tissue taken prior to immunotherapy treatment in metastatic NSCLC patients and identified a clonally expanded CD4+ T-cell population expressing a canonical cytotoxic gene program. This enabled us to develop a transcriptional signature in bulk (clinical) RNA-Seq data to detect this cell population, which robustly predicts response to immune checkpoint blockade (ICB) therapies.
Tempus found that by combining this transcriptional signature with tumor mutational burden, we could even more accurately predict response. Tempus found this to be true for the overall NSCLC population (left below) and among patients with loss of heterozygosity at the HLA-I locus (right below), a known genetic biomarker of poor response to ICB.
Figure 2. Plots showing real-world time to disease progression
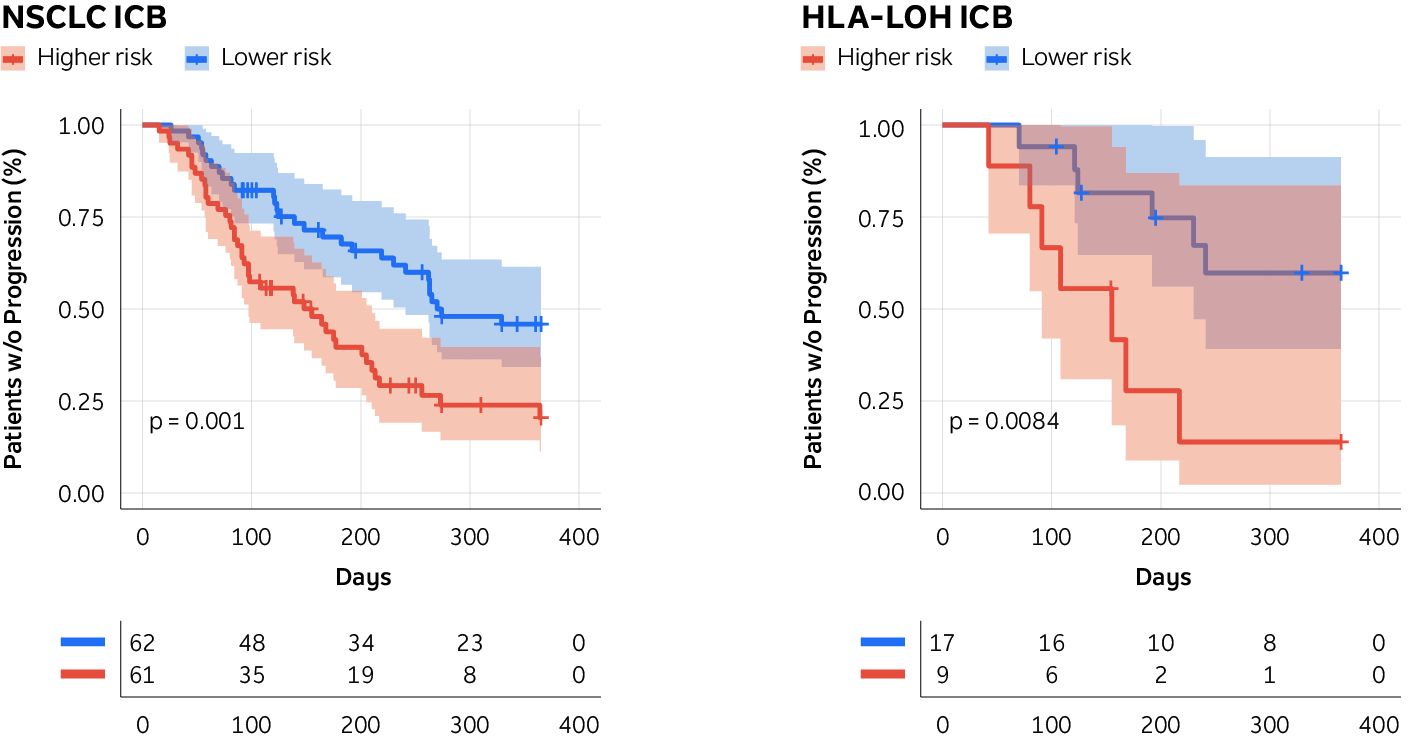
Source: Lau, D., Khare, S., Stein, M.M. et al. Integration of tumor extrinsic and intrinsic features associates with immunotherapy response in non-small cell lung cancer. Nat Commun 13, 4053 (2022). https://doi.org/10.1038/s41467-022-31769-4
These findings showcase the power of integrating multiple tumor extrinsic and intrinsic features to develop a pretreatment biomarker that can predict response to therapies available to patients today. Pretreatment biomarkers like these can help guide clinical decision making and ultimately lead to better outcomes for the patient subpopulations by limiting the use of unnecessary therapies and improving patient quality of life.
More to come
Recent drug development successes, such as with checkpoint inhibitors targeting PD-L1 and CTLA4 and therapies targeting other checkpoints like LAG3, demonstrate the value of biomarker-driven strategies and implementation.
Tempus provides a practical application that you can apply to optimize your clinical trials: Use heterogeneous data sources to examine expression profiles in precancer and specific cancer types and therapeutic settings. Research shows that “extensive molecular profiling combined with clinical data identifies personalized therapies and clinical trials for a large proportion of patients with cancer and that paired tumor-normal plus transcriptome sequencing outperforms tumor-only DNA panel testing.”
Multimodal data, like that available through the Tempus Database, offers great potential for researchers to accurately model outcomes to standard of care therapies, to uncover signatures of response or resistance to find subpopulations of unmet need, and connect potentially eligible patients to trials designed with specific biomarker strategies.
We’ve seen substantial results with our partnerships when multimodal data is applied across the drug development process. Contact us to discuss how you can use heterogeneous data sources to accelerate and de-risk your drug development programs.
Learn more

Learn more
Applications for multimodal real-world data
De-risking clinical trials and unlocking new knowledge about diseases
DOWNLOAD GUIDEStay informed
Be notified whenever Tempus publishes new and relevant research, webinars, and other resources.
Sign up-
05/24/2024
ADCs in focus: The next generation of precision medicine in oncology
Watch this engaging discussion as thought leaders explore the evolving landscape of antibody-drug conjugates (ADCs) in cancer therapeutics, including resistance mutations in first-gen ADCs, combination strategies with immune checkpoint inhibitors (ICIs), and next-gen targets and technologies. Tempus is dedicated to enhancing ADC development by offering comprehensive genomic profiling, multimodal de-identified real-world data (RWD), and AI-enabled solutions, informing target discovery and clinical trial optimization for more personalized therapies.
Watch replay -
06/07/2024
Advancing transcriptional cancer therapies with Tempus’ comprehensive life science solutions
Kronos Bio, a trailblazer in the biopharmaceutical industry, is dedicated to the discovery and development of novel therapies that target transcriptional regulation in cancer. Kronos turned to Tempus’ comprehensive suite of solutions to propel its principal asset from discovery to clinical trials.
Read moreLIVE WEBINAR
Wednesday, July 31 @ 11am CT
Integrating Omics in R&D: A new era of biomedical research
Join us as we explore how multi-omics is reshaping the research landscape. Gain valuable insights from industry leaders as we: dive into the integration of diverse omics data, from NGS to proteomics, in research; discuss the transformative role of multi-omics in understanding complex disease mechanisms; and explore how multi-omics is personalizing clinical research for precision medicine.
-
05/30/2024
Accelerating ADC Development with Tempus AI
Empowering precision and efficiency in ADC therapeutics with advanced data analytics
Read more